Introduction
Hedgehog (Hh) is a morphogen that acts a ligand for the signaling pathway critical for embryo development and adult cell maintenance. In our lab, we are researching how this pathway is regulated downstream by inhibiting and activating a transcriptional effector, Ci.
Hh signaling pathway plays an important role in embryo pattern formation, as well as adult stem cell maintenance. In humans, the mutation in the pathway can cause many different developmental abnormalities and, in adult cells, can develop cancers.
Therefore, it is important to understand how the pathway is regulated and what the components of the pathway are. The previous studies have aided in the development of effective cancer treatments.
In particular, we are currently interested in studying how the pathway is inhibited and activated, and we are trying to answer this question using the well established system in Drosophila.
Hh acts by regulating the activity of transcription factor Ci (Gli in mammals)
- With Hh
- Processing of Ci-155 is inhibited
- Ci-155 is activated by Fu kinase
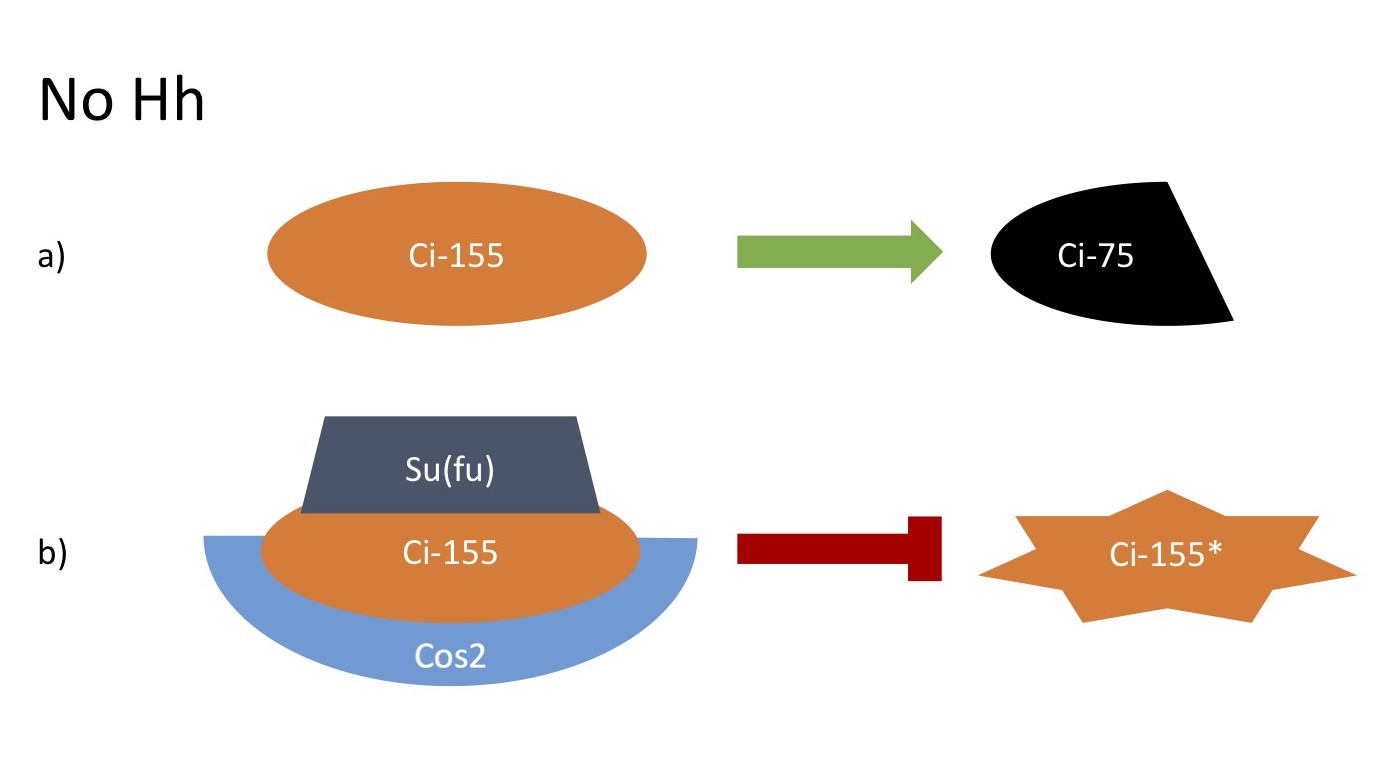
No Hh
a. Full length Ci (Ci-155) is proteolytically processed to a transcriptional repressor (Ci-75)
b. Unprocessed Ci-155 is inhibited by binding to Su(fu) and Cos2
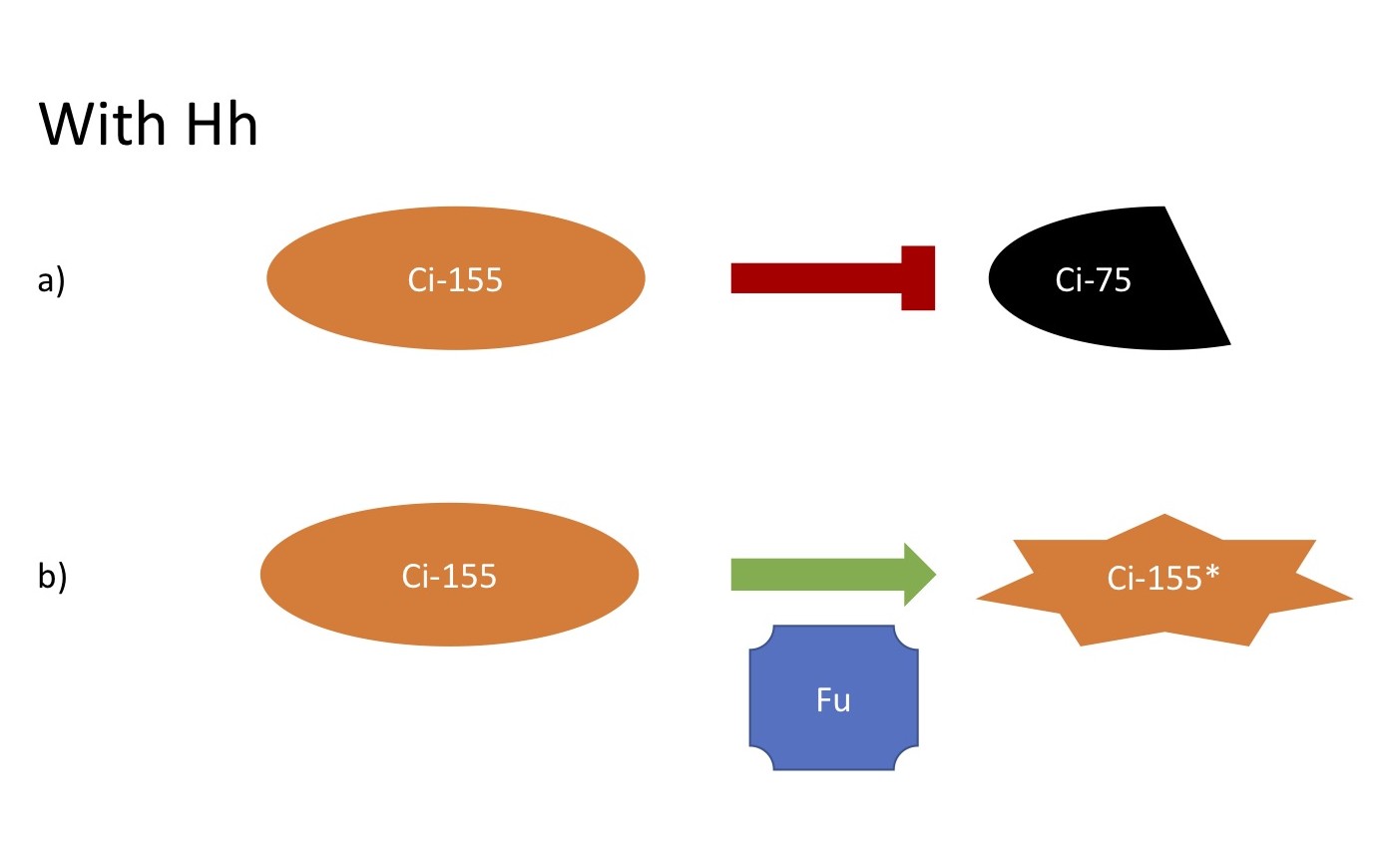
With Hh
a. Processing of Ci-155 is inhibited
b. Ci-155 is activated by Fu kinase
- How does Su(fu) inhibit Ci-155? (also applicable in mammals)
- Where does Su(fu) bind to Ci-155?
- What are the mechanisms of Su(fu) inhibition?
- Cytoplasmic anchoring
- Blocking activator
- Recruiting co-repressor
- How does Fu kinase activate Ci-155? (No Fu equivalent in mammals found yet)
- Is the direct phosphorylation of Ci necessary for activation?
How we study the Hh signaling pathway in flies.
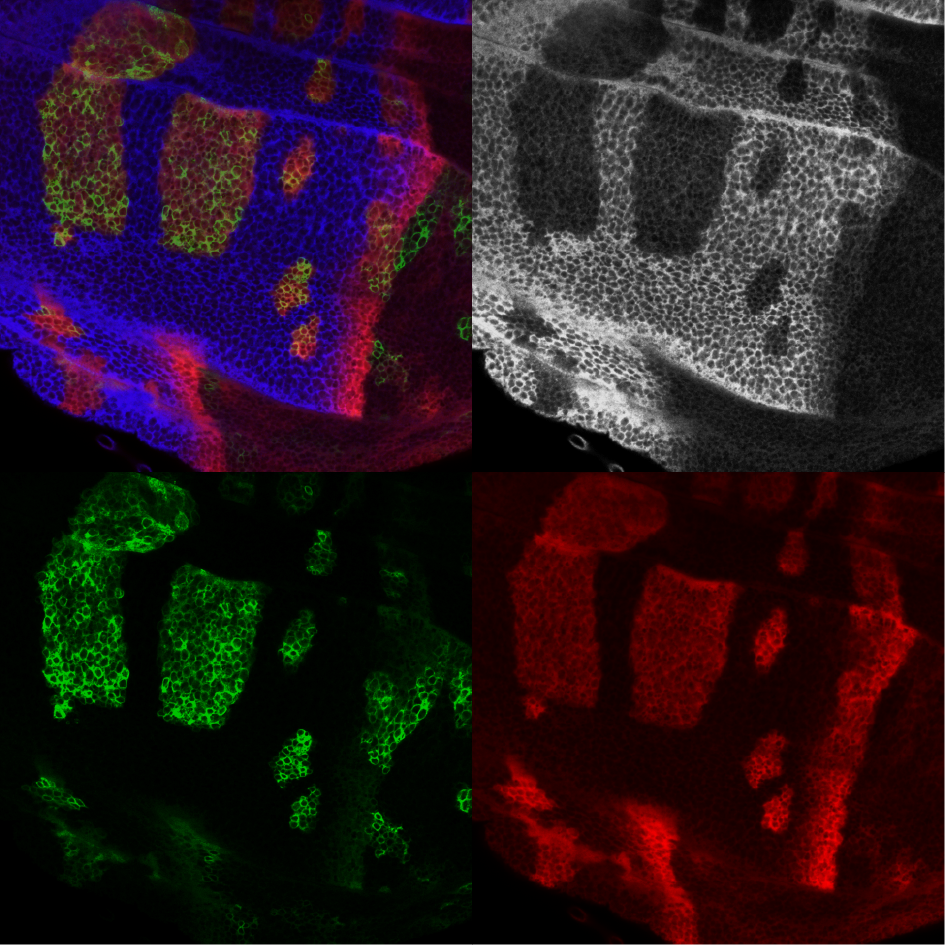
In our lab, we use larval imaginal wing disc as a system to study the mechanism of the pathway in vivo, where we can genetically manipulate each component of the pathway using CRISPR/Cas9 and monitor target gene activities under the physiological conditions via immunohistochemistry and confocal imaging.
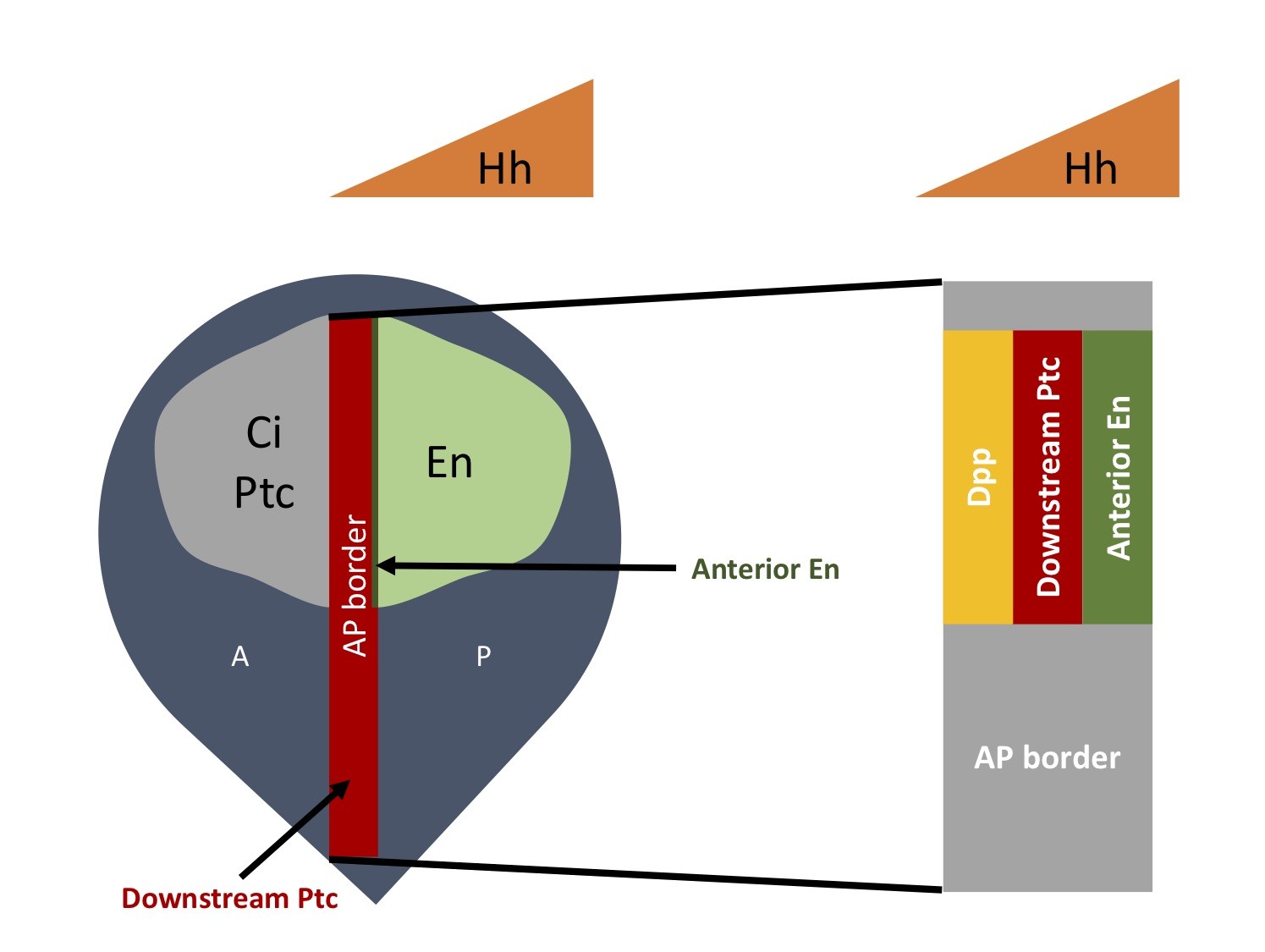
Fly wing disc is shown in navy. Hh gradient is shown in orange. The expanded region of the AP border (grey) shows the expression of downstream Hh target genes. For most of our experiments, we mainly focus on the downstream Ptc expression and Ci-155 protein level, as well as anterior En expression.
Hh signaling is critical for the normal development of fly wing, and larval wing disc provides a good system to monitor the pathway activity with clearly defined compartments and differential Hh target gene expressions.
- Fly wing disc has anterior (A) and posterior (P) compartments.
- P compartment
- Engrailed (En) is expressed and stimulates the expression of Hh
- En inhibits the expression of Patched (Ptc) and Ci in the P compartment
- A compartment
- Downstream component of the pathway, Ptc and Ci are expressed
- No Hh is expressed
- Hh travels from the P to A compartment to activate the pathway
- The most posterior A cells receiving Hh form the AP border
- Hh target genes are expressed differentially at the AP border in response to Hh signaling
- Dpp is expressed at the lowest level of Hh
- Ptc (Downstream Ptc) is expressed at the moderate level of Hh
- Anterior En is expressed at the highest level of Hh
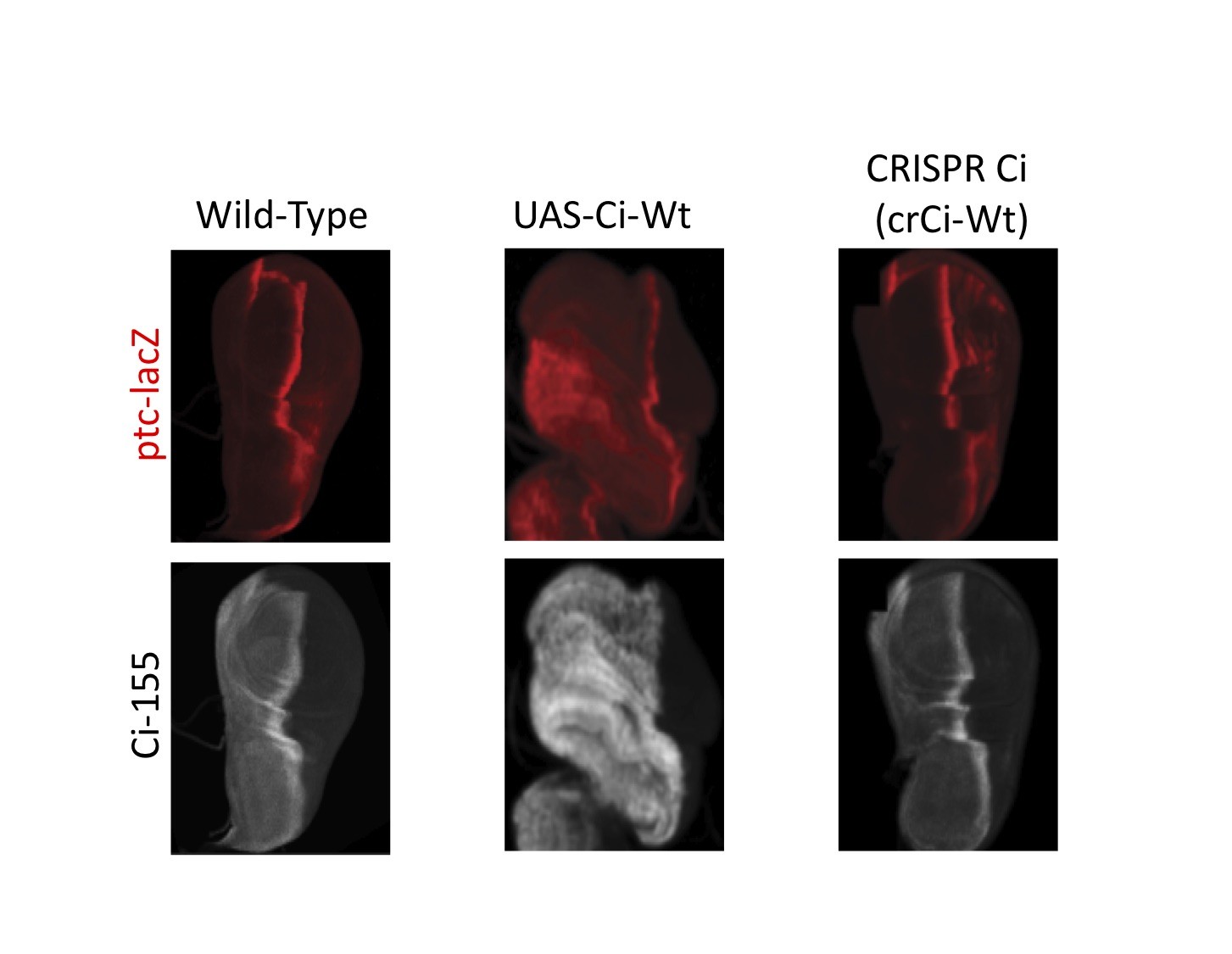
The wild type wing disc with no genetic alteration (left), wing disc where Ci is expressed using UAS system (middle), and wing disc where Ci is expressed using CRISPR/Cas9 (right) are shown here. It is clear that with UAS, Ci (grey) is over expressed and the overall shape of the wing disc is expanded compared to the wild type. CRISPR Ci looks and behaves like the Wt disc.
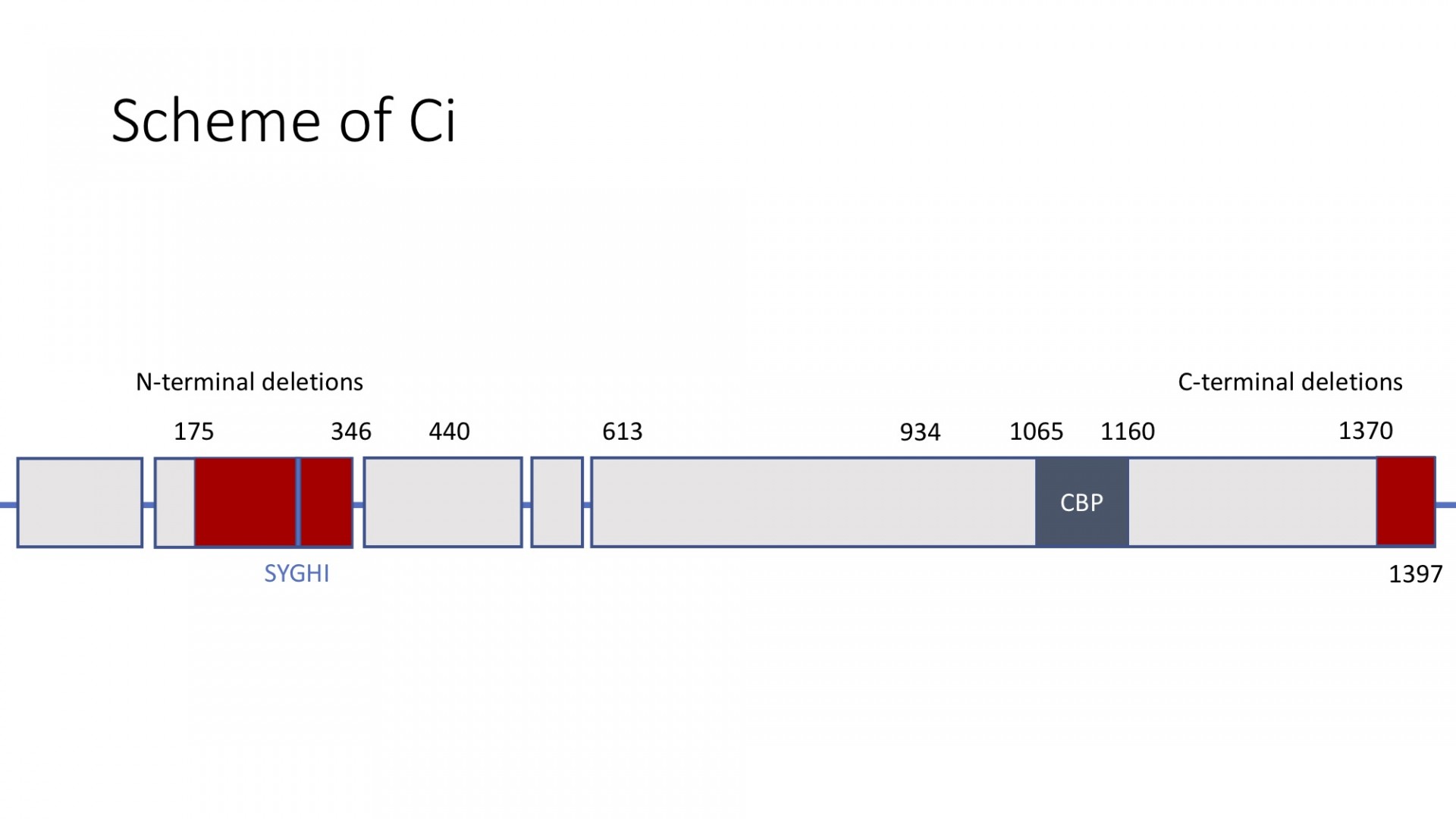
General scheme of Ci with Su(fu) binding sites shown in red
- A Ci mutation of interest is introduced via two rounds of CRISPR/Cas9 injection to create a stable male stock
- Male stock with a Ci mutation is crossed with female stocks of various Hh pathway mutant background to observe the effects of the mutations
- Female stocks include: Wt background, Su(fu) mutant background, Fu mutant background, Cos2 clones and constitutively active Fu clones
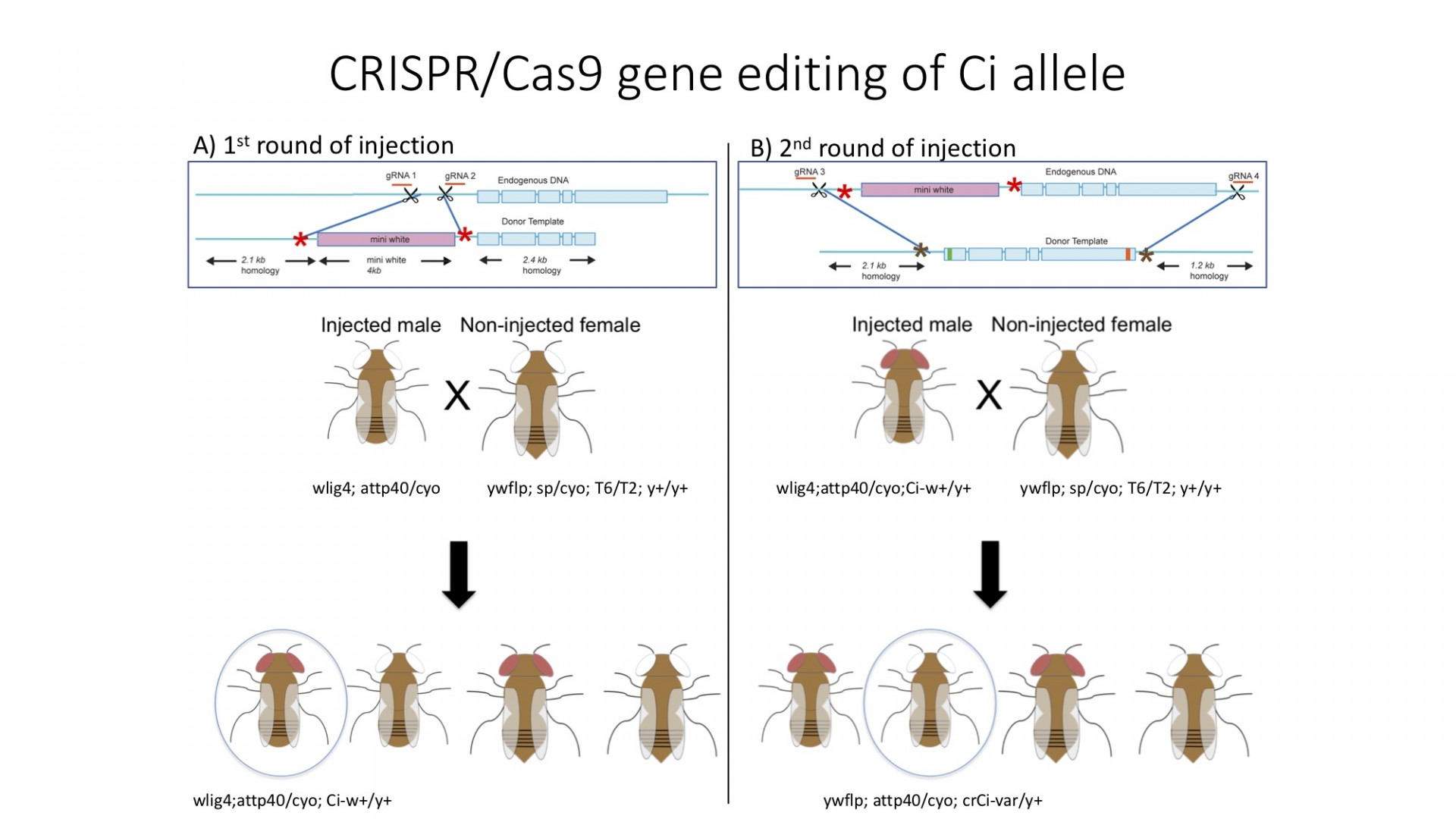
- Mini white gene, responsible for the eye color of the fly is introduced to the first intron of ci gene in the first round of injection, and offspring with the eye color is selected to create a basal injection stock.
- Ci construct with the mutation of interest replaces the whole region of Ci between the first intron and the 3' UTR, which will also replace the mini white gene introduced in the first round of injection. Offspring with no eye color is selected to create a stable stock of males for the future experiments.